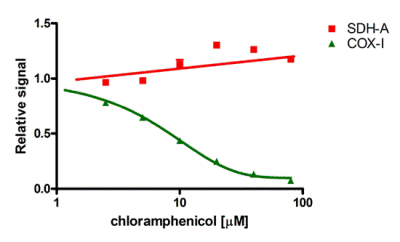
Inhibition of mitochondrial biogenesis by chloramphenicol The IC50 of a drug's effect on mitochondrial protein translation can be determined quickly using the MitoBiogenesis™ In-Cell ELISA Kit. In this example, cells were seeded at 6000 cells/well, allowed to grow for 3 cell doublings in a drug dilution series and then the relative amounts of COX-I, and SDH-A were measured in each well. Chloramphenicol inhibits mtDNA-encoded COX-I protein synthesis relative to nuclear DNA-encoded SDH-A protein synthesis by 50% at 8.1 µM.
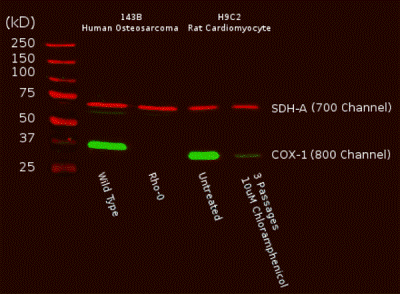
Antibody specificity demonstrated by Western Blot. A Western blot of total cell protein (10 µg) from human or rat cultured cells was probed with the primary and secondary antibodies and scanned with a LI-COR® Odyssey® imager. The two mitochondrial proteins targeted by the two primary mAbs were labeled and visualized specifically despite the presence of thousands of other proteins. Furthermore, reduction of mtDNA levels in human Rho0 (mtDNA-depleted) cells, or inhibition of mitochondrial protein translation by chloramphenicol in rat cells both result in specific reduction of COX-I protein while nuclear DNA-encoded SDH-A is unaffected.
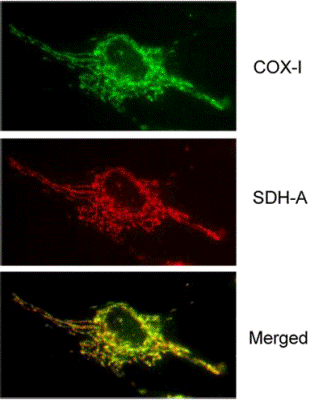
Antibody specificity demonstrated by immunocytochemistry. Two-color immunocytochemical labeling of cultured cells with the two MS643 primary monoclonal antibodies specific for COX-I and SDH-A. The two antibodies exhibit striking and specific co-localization in the mitochondria, consistent with the known mitochondrial expression of both proteins.
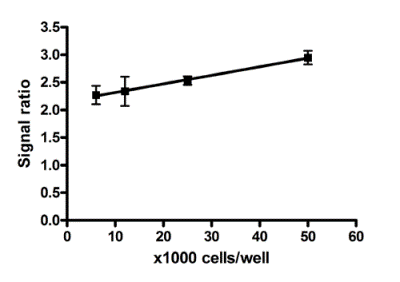
Quantitative measurement of the COX-I/SDH-A protein expression ratio. At all cell concentrations, a consistent ratio of mtDNA-encoded protein expression (COX-I) to nuclear DNA-encoded mitochondrial protein expression (SDH-A) is observed in untreated cells. Therefore, normalizing COX-I levels to SDH-A levels simplifies data analysis and eliminates the need to perform all tests at the same cell concentration.
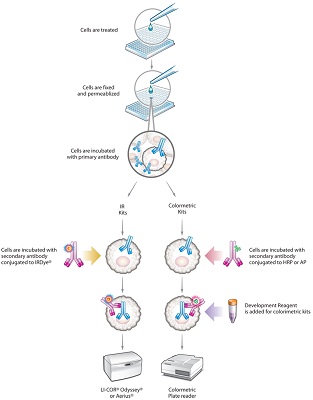
Cells are grown to ~80% confluency in a 96- or 384-well plate, a drug/other treatment is applied to stimulate a cellular response. The cells are then fixed and permeabilized, effectively "freezing" them. Primary antibodies are then added which bind to their intended targets within the mitochondria or other subcellular compartment. After incubation, the unbound primary antibodies are washed away and secondary antibodies are added. These secondaries are conjugated to either IRDyes® or to an enzyme label (HRP or AP) for the colorimetric versions of the assays. Unbound secondaries are washed away, reaction buffer is added for the colorimetric assays, and the signal is read on a suitable instrument for the kit type.» In-cell ELISA diagram in PDF format




