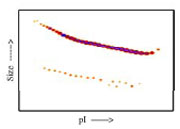
Densitometry of protein isoforms visualised by 2-DE. The triangle indicates the theoretical MW and pI of the protein.
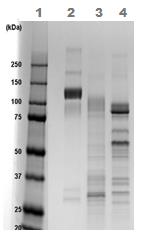
1D SDS-PAGE of ab83682 before and after treatment with glycosidases to remove oligosaccharides. Lane 1 MW markers; Lane 2 ab83682; Lane 3 ab83682 treated with PNGase F to remove potential N-linked glycans; Lane 4 ab83682 treated with a glycosidase cocktail to remove potential N- and O-linked glycans. Approximately 5 µg of protein was loaded per lane. Drop in MW after treatment with PNGase F indicates presence of N-linked glycans. A further drop in MW after treatment with the glycosidase cocktail indicates the presence of O-linked glycans. Additional bands in lane 3 and lane 4 are glycosidase enzymes.
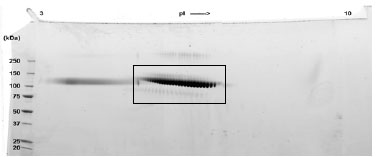
A sample of ab83682 without carrier protein was reduced and alkylated and focused on a 3-10 IPG strip then run on a 4-20% Tris-HCl 2D gel. Approximately 40 µg of protein was load; Gel was stained using Deep Purple™.


