| Gene Symbol |
Clock
|
| Entrez Gene |
12753
|
| Alt Symbol |
5330400M04Rik, KAT13D
|
| Species |
Mouse
|
| Gene Type |
protein-coding
|
| Description |
circadian locomotor output cycles kaput
|
| Other Description |
circadian locomoter output cycles protein kaput
|
| Swissprots |
O08785
|
| Accessions |
AAD30565 EDL37895 EDL37896 O08785 AB221635 AF000998 AAC53200 AK030352 AK040337 BAC30568 AK041303 BAC30897 AK129118 BAC97928 AK132842 AK136367 BAE22949 AK140803 AK144118 AK158765 AY118227 AAM78583 BC129886 BC148468 AAI48469 BC156741 AAI56742 BX526669 XM_006534744 XP_006534807 XM_006534745 XP_006534808 XM_011249400 XP_011247702 XM_011249401 XP_011247703 XM_011249402 XP_011247704 XM_011249403 XP_011247705 NM_001289826 NP_001276755 NM_001305222 NP_001292151 NM_007715 NP_031741
|
| Function |
Transcriptional activator which forms a core component of the circadian clock. The circadian clock, an internal time- keeping system, regulates various physiological processes through the generation of approximately 24 hour circadian rhythms in gene expression, which are translated into rhythms in metabolism and behavior. It is derived from the Latin roots 'circa' (about) and 'diem' (day) and acts as an important regulator of a wide array of physiological functions including metabolism, sleep, body temperature, blood pressure, endocrine, immune, cardiovascular, and renal function. Consists of two major components: the central clock, residing in the suprachiasmatic nucleus (SCN) of the brain, and the peripheral clocks that are present in nearly every tissue and organ system. Both the central and peripheral clocks can be reset by environmental cues, also known as Zeitgebers (German for 'timegivers'). The predominant Zeitgeber for the central clock is light, which is sensed by retina and
|
| Subcellular Location |
Nucleus {ECO:0000269|PubMed:11779462, ECO:0000269|PubMed:12897057, ECO:0000269|PubMed:16980631, ECO:0000269|PubMed:17310242, ECO:0000269|PubMed:18662546, ECO:0000269|PubMed:19414601}. Cytoplasm {ECO:0000269|PubMed:12897057, ECO:0000269|PubMed:16980631}. Note=Localizes to sites of DNA damage in a H2AX-independent manner (By similarity). Shuffling between the cytoplasm and the nucleus is under circadian regulation and is ARNTL/BMAL1-dependent. Phosphorylated form located in the nucleus predominantly between CT12 and CT21. Nonphosphorylated form found only in the cytoplasm. Sequestered to the cytoplasm in the presence of ID2. {ECO:0000250|UniProtKB:O15516, ECO:0000269|PubMed:11779462, ECO:0000269|PubMed:16980631, ECO:0000269|PubMed:20861012}.
|
| Tissue Specificity |
Expressed equally in brain, eye, testes, ovaries, liver, heart, lung, kidney. In the brain, expression is abundant in the suprachiasmatic nuclei (SCN), in the pyriform cortex, and in the hippocampus. Low expression throughout the rest of the brain. Expression does not appear to undergo circadian oscillations. {ECO:0000269|PubMed:22900038, ECO:0000269|PubMed:24154698, ECO:0000269|PubMed:9160755}.
|
| Top Pathways |
Circadian rhythm, Herpes simplex infection, Dopaminergic synapse
|
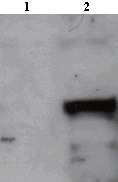
![Western Blot: CLOCK Antibody [NBP1-30326] - Analysis of CLOCK in Sol8 (mouse embryo fibroblast) cell extracts using NBP1-30326.](http://www.bioprodhub.com/system/product_images/ab_products/5/sub_1/21517_CLOCK-Antibody-Western-Blot-NBP1-30326-img0007.jpg)