| Gene Symbol |
SMARCAD1
|
| Entrez Gene |
56916
|
| Alt Symbol |
ADERM, ETL1, HEL1
|
| Species |
Human
|
| Gene Type |
protein-coding
|
| Description |
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1
|
| Other Description |
ATP-dependent helicase 1|SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1
|
| Swissprots |
Q05D56 Q96SX1 Q9H4L7 B7Z799 Q9H017 Q9ULU7 Q9H860 Q9NPU9
|
| Accessions |
EAX06049 EAX06050 EAX06051 Q9H4L7 AB032948 BAA86436 AK023990 BAB14759 AK025302 AK027490 BAB55150 AK301668 BAH13535 AL359929 CAB95769 AL512768 CAC21685 AL704040 AY008271 AAG16639 BC017953 AAH17953 BC041800 BC045534 AAH45534 DA798320 DB207879 EU446826 ABZ92355 EU832178 ACE87528 EU832271 ACE86841 XR_938765 XR_938766 NM_001128429 NP_001121901 NM_001128430 NP_001121902 NM_001254949 NP_001241878 NM_020159 NP_064544 NR_045644
|
| Function |
DNA helicase that possesses intrinsic ATP-dependent nucleosome-remodeling activity and is both required for DNA repair and heterochromatin organization. Promotes DNA end resection of double-strand breaks (DSBs) following DNA damage: probably acts by weakening histone DNA interactions in nucleosomes flanking DSBs. Required for the restoration of heterochromatin organization after replication. Acts at replication sites to facilitate the maintenance of heterochromatin by directing H3 and H4 histones deacetylation, H3 'Lys-9' trimethylation (H3K9me3) and restoration of silencing. {ECO:0000269|PubMed:21549307, ECO:0000269|PubMed:22960744}.
|
| Subcellular Location |
Nucleus. Chromosome. Note=Colocalizes with PCNA at replication forks during S phase. Recruited to double- strand breaks (DSBs) sites of DNA damage.
|
| Tissue Specificity |
Isoform 1 is expressed ubiquitously. Isoform 3 is expressed mainly in skin, fibroblasts, keratinocytes and esophagus. {ECO:0000269|PubMed:11031099, ECO:0000269|PubMed:21820097}.
|
| Top Pathways |
Signaling pathways regulating pluripotency of stem cells
|

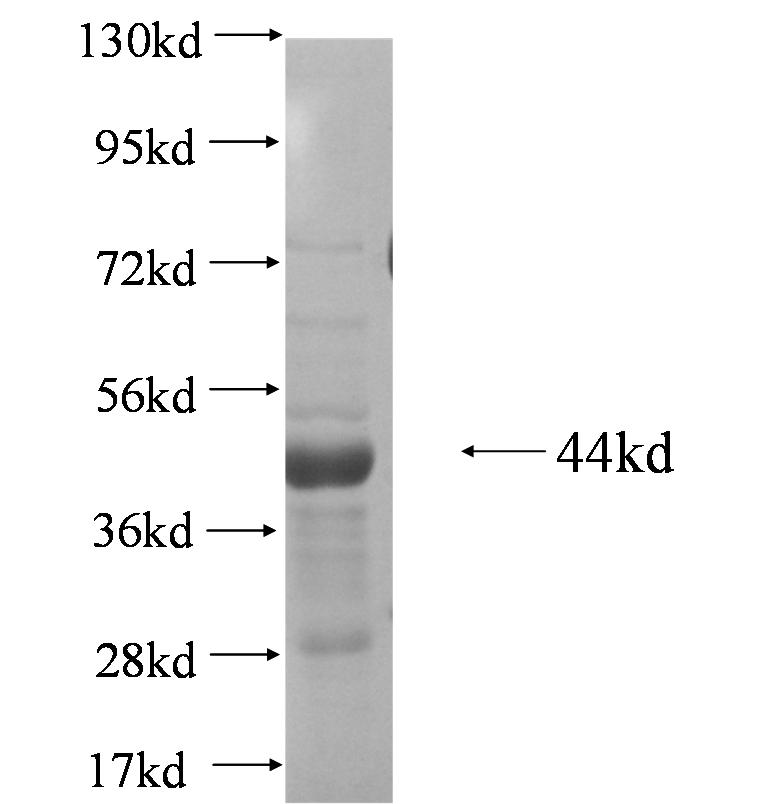
![SDS-Page: SMARCAD1 Recombinant Protein [H00056916-P01]](http://www.bioprodhub.com/system/product_images/pp_products/5/sub_1/20900_SMARCAD1-Recombinant-Protein-SDS-Page-H00056916-P01-img0001.jpg)