| Gene Symbol |
STUB1
|
| Entrez Gene |
10273
|
| Alt Symbol |
CHIP, HSPABP2, NY-CO-7, SCAR16, SDCCAG7, UBOX1
|
| Species |
Human
|
| Gene Type |
protein-coding
|
| Description |
STIP1 homology and U-box containing protein 1, E3 ubiquitin protein ligase
|
| Other Description |
CLL-associated antigen KW-8|E3 ubiquitin-protein ligase CHIP|antigen NY-CO-7|carboxy terminus of Hsp70-interacting protein|heat shock protein A binding protein 2 (c-terminal)|serologically defined colon cancer antigen 7
|
| Swissprots |
O60526 Q969U2 A2IDB9 Q9HBT1 Q9UNE7
|
| Accessions |
AAK61242 CAV30716 EAW85756 EAW85757 EAW85758 Q9UNE7 AF039689 AAC18038 AF129085 AAD33400 AF217968 AAG17211 AF432221 AAL99927 BC007545 AAH07545 BC017178 AAH17178 BC022788 AAH22788 BC063617 AAH63617 CB145452 DQ893185 ABM84111 DQ896502 ABM87501 NM_001293197 NP_001280126 NM_005861 NP_005852
|
| Function |
E3 ubiquitin-protein ligase which targets misfolded chaperone substrates towards proteasomal degradation. Collaborates with ATXN3 in the degradation of misfolded chaperone substrates: ATXN3 restricting the length of ubiquitin chain attached to STUB1/CHIP substrates and preventing further chain extension. Ubiquitinates NOS1 in concert with Hsp70 and Hsp40. Modulates the activity of several chaperone complexes, including Hsp70, Hsc70 and Hsp90. Mediates transfer of non-canonical short ubiquitin chains to HSPA8 that have no effect on HSPA8 degradation. Mediates polyubiquitination of DNA polymerase beta (POLB) at 'Lys-41', 'Lys-61' and 'Lys-81', thereby playing a role in base-excision repair: catalyzes polyubiquitination by amplifying the HUWE1/ARF- BP1-dependent monoubiquitination and leading to POLB-degradation by the proteasome. Mediates polyubiquitination of CYP3A4. Ubiquitinates EPHA2 and may regulate the receptor stability and activity through proteasomal degradation. Negatively regu
|
| Subcellular Location |
Cytoplasm {ECO:0000269|PubMed:10330192, ECO:0000269|PubMed:23973223}. Nucleus {ECO:0000269|PubMed:23973223}. Note=Translocates to the nucleus in response to inflammatory signals in regulatory T-cells (Treg). {ECO:0000269|PubMed:23973223}.
|
| Tissue Specificity |
Highly expressed in skeletal muscle, heart, pancreas, brain and placenta. Detected in kidney, liver and lung. {ECO:0000269|PubMed:10330192, ECO:0000269|PubMed:11435423}.
|
| Top Pathways |
Ubiquitin mediated proteolysis, Protein processing in endoplasmic reticulum
|

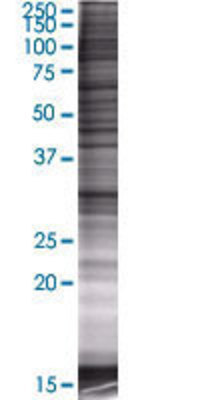
![Western Blot: CHIP Overexpression Lysate (Adult Normal) [NBP2-07696] Left-Empty vector transfected control cell lysate (HEK293 cell lysate); Right -Over-expression Lysate for CHIP.](http://www.bioprodhub.com/system/product_images/cel_products/5/sub_2/8562_CHIP%2520Overexpression%2520Lysate%2520(Adult%2520Normal)-Western%2520Blot-NBP2-07696-img0001.jpg)