| Gene Symbol |
FBXO6
|
| Entrez Gene |
26270
|
| Alt Symbol |
FBG2, FBS2, FBX6, Fbx6b
|
| Species |
Human
|
| Gene Type |
protein-coding
|
| Description |
F-box protein 6
|
| Other Description |
F-box only protein 6|F-box protein FBG2|F-box protein Fbx6|F-box protein that recognizes sugar chains 2|F-box/G-domain protein 2
|
| Swissprots |
B2RC88 Q9UKT3 B1AK42 Q9NRD1
|
| Accessions |
EAW71696 Q9NRD1 AF129536 AAF04470 AF233223 AAF67153 AK314989 BAG37485 BC020880 AAH20880 HQ447671 ADQ32156 XM_005263448 XP_005263505 XM_005263449 XP_005263506 XM_005263451 XP_005263508 XM_006710571 XP_006710634 XM_011541232 XP_011539534 NM_018438 NP_060908
|
| Function |
Substrate-recognition component of some SCF (SKP1-CUL1- F-box protein)-type E3 ubiquitin ligase complexes. Involved in endoplasmic reticulum-associated degradation pathway (ERAD) for misfolded lumenal proteins by recognizing and binding sugar chains on unfolded glycoproteins that are retrotranlocated into the cytosol and promoting their ubiquitination and subsequent degradation. Able to recognize and bind denatured glycoproteins, which are modified with not only high-mannose but also complex- type oligosaccharides. Also recognizes sulfated glycans. Also involved in DNA damage response by specifically recognizing activated CHEK1 (phosphorylated on 'Ser-345'), promoting its ubiquitination and degradation. Ubiquitination of CHEK1 is required to insure that activated CHEK1 does not accumulate as cells progress through S phase, or when replication forks encounter transient impediments during normal DNA replication. {ECO:0000269|PubMed:18203720, ECO:0000269|PubMed:19716789}.
|
| Subcellular Location |
Cytoplasm {ECO:0000269|PubMed:19716789}.
|
| Top Pathways |
Protein processing in endoplasmic reticulum
|
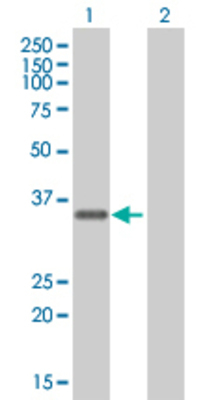
![Western Blot: FBXO6 Overexpression Lysate (Adult Normal) [NBL1-10640] Left-Empty vector transfected control cell lysate (HEK293 cell lysate); Right -Over-expression Lysate for FBXO6.](http://www.bioprodhub.com/system/product_images/cel_products/5/sub_2/8785_FBXO6%2520Overexpression%2520Lysate%2520(Adult%2520Normal)-Western%2520Blot-NBL1-10640-img0002.jpg)