| Gene Symbol |
SIRT3
|
| Entrez Gene |
23410
|
| Alt Symbol |
SIR2L3
|
| Species |
Human
|
| Gene Type |
protein-coding
|
| Description |
sirtuin 3
|
| Other Description |
NAD-dependent deacetylase sirtuin-3, mitochondrial|NAD-dependent protein deacetylase sirtuin-3, mitochondrial|SIR2-like protein 3|mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase|regulatory protein SIR2 homolog 3|silent mating type information regulation 2, S.cerevisiae, homolog 3|sir2-like 3|sirtuin type 3
|
| Swissprots |
Q9Y6E8 B7Z5U6 Q9NTG7
|
| Accessions |
CAL00156 CAL00157 CAL48199 CAN36293 EAW61239 EAW61240 EAW61241 EAW61242 EAW61243 Q9NTG7 AA279020 AF083108 AAD40851 AI091200 AK074992 AK298572 BAH12816 AK299009 BAH12927 AK299438 BAH13032 AK301558 BAH13513 AK301979 BAH13600 AK308681 AK316175 BAH14546 AL137276 CAB70674 AL535769 BC001042 AAH01042 BI755839 BM709066 BM973763 DR762907 XM_005252835 XP_005252892 XM_011519956 XP_011518258 XM_011519957 XP_011518259 NM_001017524 NP_001017524 NM_012239 NP_036371
|
| Function |
NAD-dependent protein deacetylase. Activates or deactivates mitochondrial target proteins by deacetylating key lysine residues. Known targets include ACSS1, IDH, GDH, SOD2, PDHA1, LCAD, SDHA and the ATP synthase subunit ATP5O. Contributes to the regulation of the cellular energy metabolism. Important for regulating tissue-specific ATP levels. {ECO:0000269|PubMed:16788062, ECO:0000269|PubMed:18680753, ECO:0000269|PubMed:18794531, ECO:0000269|PubMed:19535340, ECO:0000269|PubMed:24121500, ECO:0000269|PubMed:24252090}.
|
| Subcellular Location |
Mitochondrion matrix {ECO:0000269|PubMed:12186850, ECO:0000269|PubMed:12374852, ECO:0000269|PubMed:16079181, ECO:0000269|PubMed:18215119}.
|
| Tissue Specificity |
Widely expressed. {ECO:0000269|PubMed:10381378, ECO:0000269|PubMed:12374852}.
|
| Top Pathways |
Central carbon metabolism in cancer
|
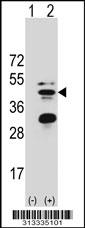
![Western Blot: SIRT3 Overexpression Lysate (Adult Normal) [NBL1-15975] Left-Empty vector transfected control cell lysate (HEK293 cell lysate); Right -Over-expression Lysate for SIRT3.](http://www.bioprodhub.com/system/product_images/cel_products/5/sub_2/4596_SIRT3%2520Overexpression%2520Lysate%2520(Adult%2520Normal)-Western%2520Blot-NBL1-15975-img0002.jpg)
![Western Blot: SIRT3 Overexpression Lysate (Adult Normal) [NBL1-15976] Left-Empty vector transfected control cell lysate (HEK293 cell lysate); Right -Over-expression Lysate for SIRT3.](http://www.bioprodhub.com/system/product_images/cel_products/5/sub_2/929_SIRT3%2520Overexpression%2520Lysate%2520(Adult%2520Normal)-Western%2520Blot-NBL1-15976-img0002.jpg)